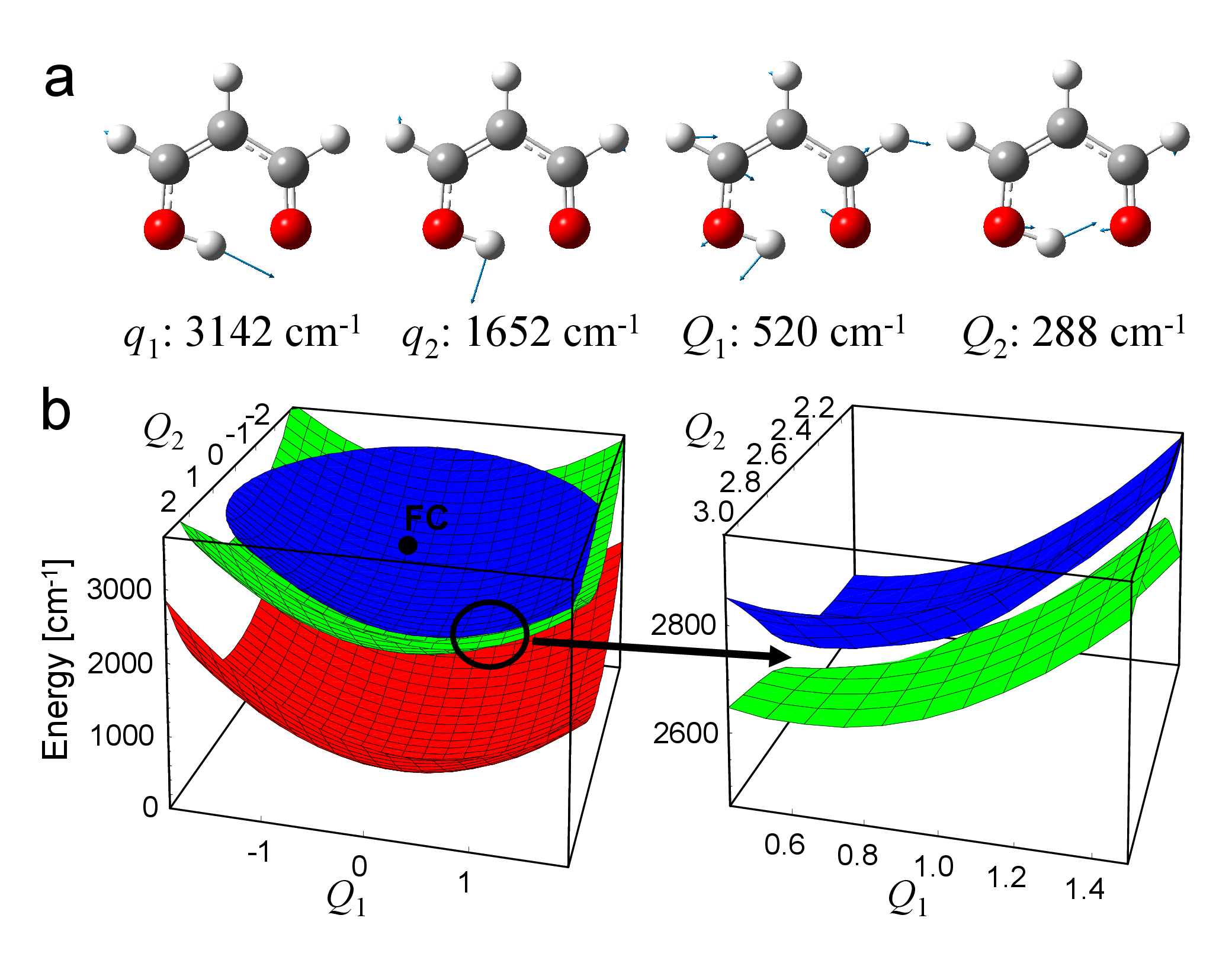
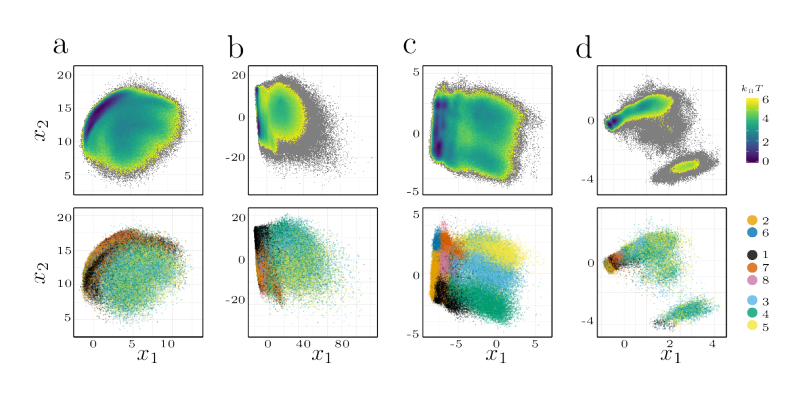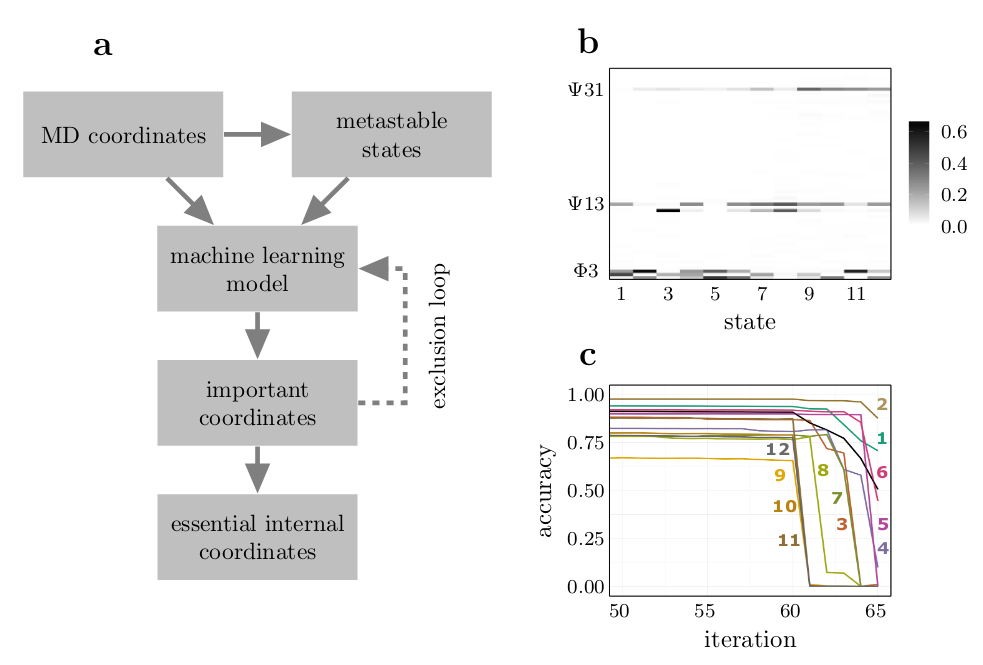Our Research
Here are some (but not all) of our research interests. For more information, please visit the publications page.
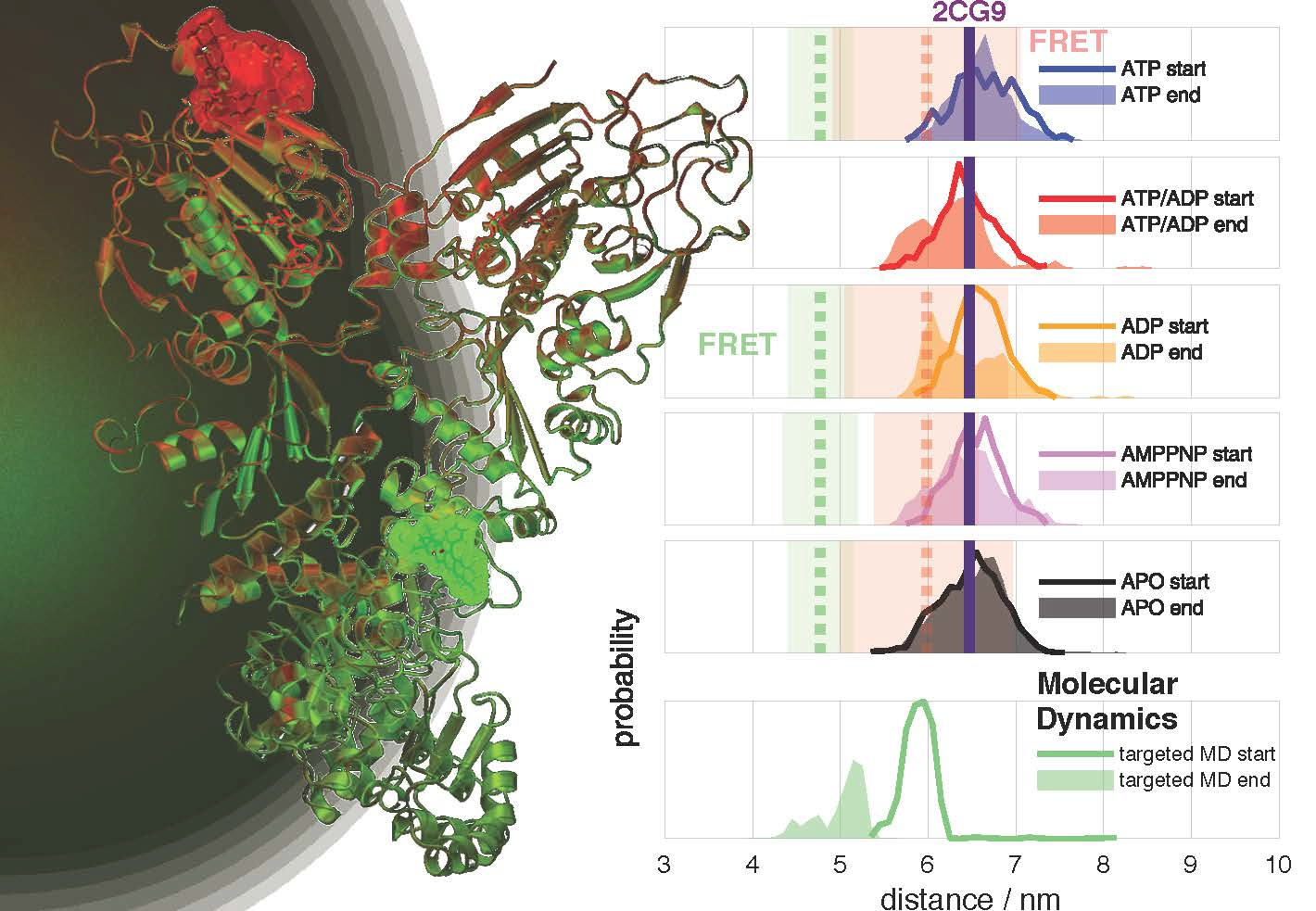
Combining single molecule FRET measurements and MD simulations to elucidate hierachical dynamics in allostery
We report on a study that combines advanced fluorescence methods with molecular dynamics (MD) simulations to cover timescales from nanoseconds to milliseconds for a large protein. This allows us to delineate how ATP hydrolysis in a protein causes allosteric changes at a distant protein binding site, using the chaperone Hsp90 as test system. The allosteric process occurs via hierarchical dynamics involving timescales from nano- to milliseconds and length scales from Ångstroms to several nanometers. We find that hydrolysis of one ATP is coupled to a conformational change of Arg380, which in turn passes structural information via the large M-domain α-helix to the whole protein. The resulting structural asymmetry in Hsp90 leads to the collapse of a central folding substrate binding site, causing the formation of a novel collapsed state (closed state B) that we characterise structurally. We presume that similar hierarchical mechanisms are fundamental for information transfer induced by ATP hydrolysis through many other proteins.
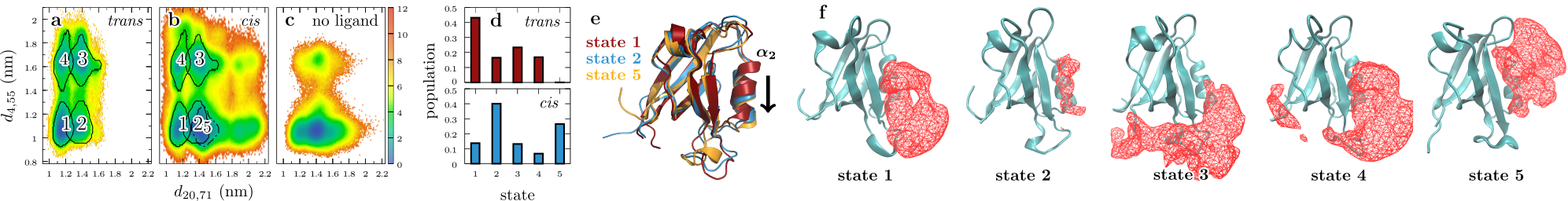
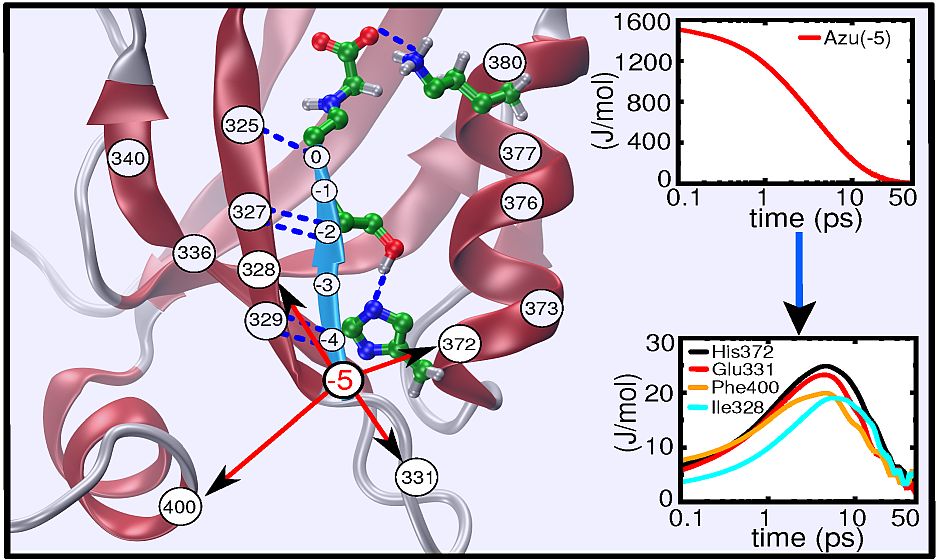
Real-time observation of ligand-induced allosteric transitions
While allostery is of paramount importance for protein regulation, the
underlying dynamical process of ligand (un)binding at one site, resulting time
evolution of the protein structure, and change of the binding affinity at a
remote site is not well understood. Here the ligand-induced conformational
transition in a widely studied model system of allostery, the PDZ2 domain, is
investigated by transient infrared spectroscopy accompanied by molecular
dynamics simulations. To this end, an azobenzene derived photoswitch is linked
to a peptide ligand in a way that its binding affinity to the PDZ2 domain
changes upon switching, thus initiating an allosteric transition in the PDZ2
domain protein. The subsequent response of the protein, covering four decades
of time ranging from ~1ns to ~10μs, can be rationalize by a
remodelling of its rugged free energy landscape, with very subtle shifts in the
populations of a small number of structurally well defined states. It is
proposed that structurally and dynamically driven allostery, often discussed as
limiting scenarios of allosteric communication, actually go hand-in-hand,
allowing the protein to adapt its free energy landscape to incoming signals.
•
Real-time observation of ligand-induced allosteric transitions in a PDZ domain,
PNAS 117, 26031 (2020), arXiv:2008.01625
Nonequilibrium MD simulations of a proteins allosteric communication
Allostery represents a fundamental mechanism of biological regulation that is mediated via long-range communication between distant protein sites. Although little is known about the underlying dynamical process, recent time-resolved infrared spectroscopy experiments on a photoswitchable PDZ domain (PDZ2S) have indicated that the allosteric transition occurs on multiple timescales. Here, using extensive nonequilibrium molecular dynamics simulations, a time-dependent picture of the allosteric communication in PDZ2S is developed. The simulations reveal that allostery amounts to the propagation of structural and dynamical changes that are genuinely nonlinear and can occur in a nonlocal fashion. A dynamic network model is constructed that illustrates the hierarchy and exceeding structural heterogeneity of the process. In compelling agreement with experiment, three physically distinct phases of the time evolution are identified, describing elastic response (≲0.1 ns), inelastic reorganization (∼100 ns), and structural relaxation (≳1μs). Issues such as the similarity to downhill folding as well as the interpretation of allosteric pathways are discussed.
•
Time-resolved observation of protein allosteric communication,
PNAS 114, 33, E6804 (2017)
Energy transport through molecular systems has recently received considerable interest,
in particular, due to its importance for molecular electronics and the functioning of
biological systems. In experiment, energy is usually induced into the molecule by
photoexcitation of a chromophore or by pumping a localized infrared vibration.
This triggers an cascade of dynamical processes including
- the femtosecond vibrational energy redistribution from localized high-frequency modes to delocalized low-frequency modes,
- transport of energy through the molecule, and
- the cooling the molecules due to heat transfer to the solvent.
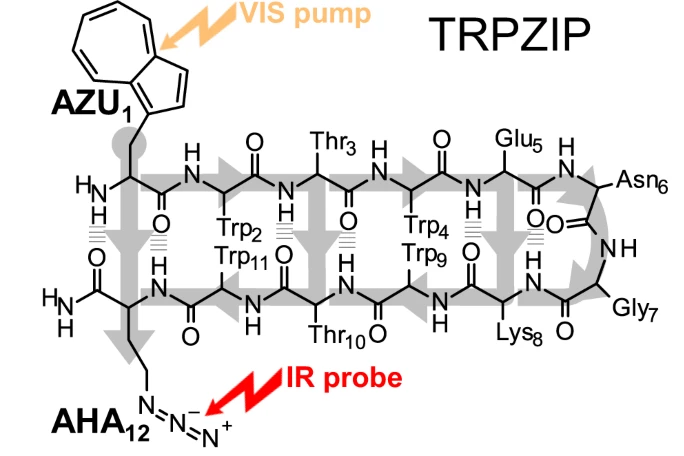
Through Bonds or Contacts? Mapping Protein Vibrational Energy Transfer Using Non-canonical Amino Acids
We examine vibrational energy flow in dehydrated and hydrated villin headpiece subdomain HP36 by
master equation simulations. Transition rates used in the simulations are obtained from communication
maps calculated for HP36. In addition to energy flow along the main chain, we identify pathways
for energy transport in HP36 via hydrogen bonding between residues quite far in sequence space. The
results of the master equation simulations compare well with all-atom non-equilibrium simulations
to about 1 ps following initial excitation of the protein, and quite well at long times, though for some
residues we observe deviations between the master equation and all-atom simulations at intermediate
times from about 1–10 ps. Those deviations are less noticeable for hydrated than dehydrated HP36
due to energy flow into the water.
•
Through Bonds or Contacts? Mapping Protein Vibrational Energy Transfer Using Non-canonical Amino Acids
Nat. Commun. 12, 3284 (2021)
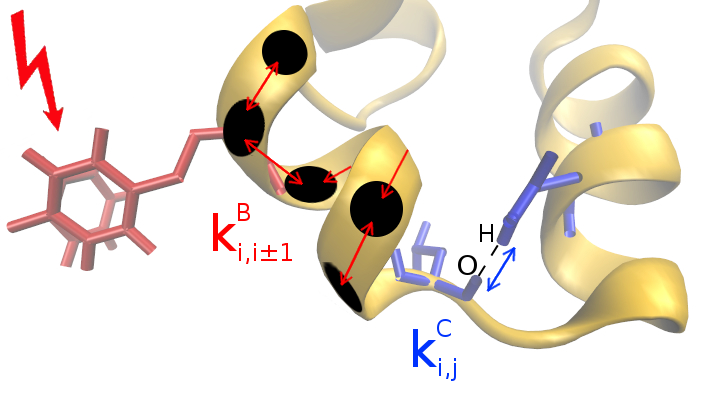
Scaling Rules for Energy Transport
We examine vibrational energy flow in dehydrated and hydrated villin headpiece subdomain HP36 by
master equation simulations. Transition rates used in the simulations are obtained from communication
maps calculated for HP36. In addition to energy flow along the main chain, we identify pathways
for energy transport in HP36 via hydrogen bonding between residues quite far in sequence space. The
results of the master equation simulations compare well with all-atom non-equilibrium simulations
to about 1 ps following initial excitation of the protein, and quite well at long times, though for some
residues we observe deviations between the master equation and all-atom simulations at intermediate
times from about 1–10 ps. Those deviations are less noticeable for hydrated than dehydrated HP36
due to energy flow into the water.
•
Scaling Rules for Vibrational Energy Transport in Globular Proteins
J. Phys. Chem. Lett. 7, 25 (2016)
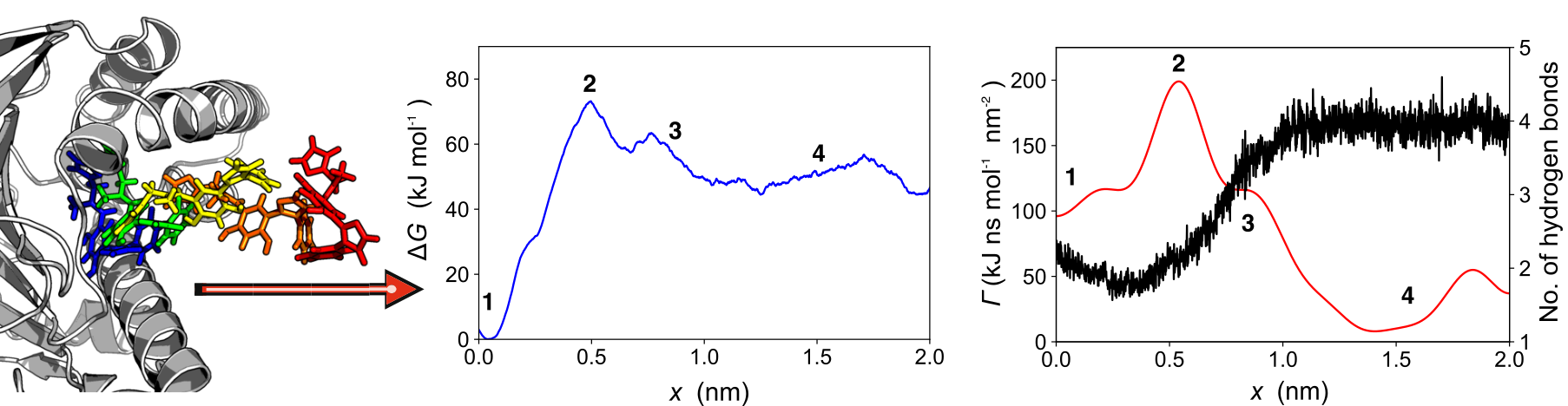
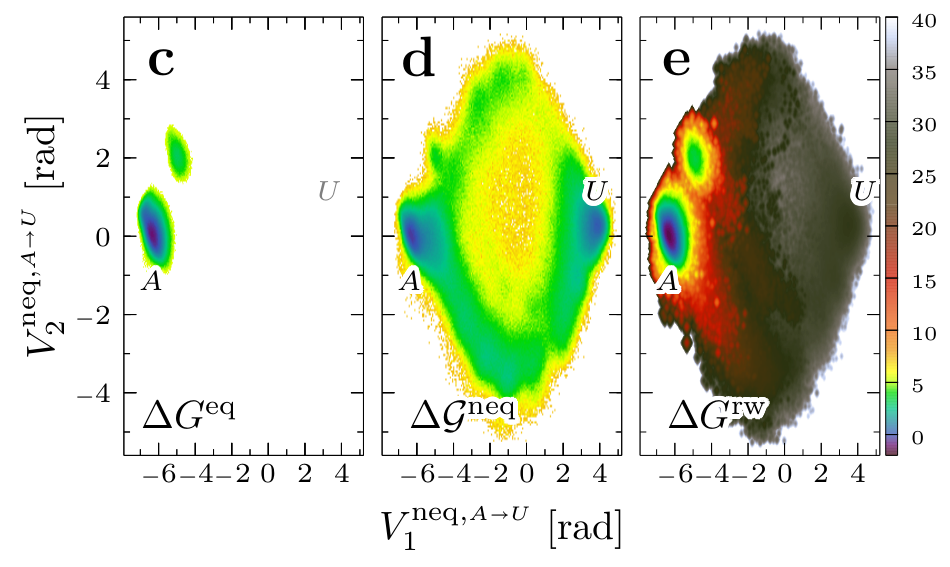
While common molecular dynamics simulation methods are capable to simulate molecular processes on timescales up to milliseconds,
the biologically relevant range of timescales reaches from microseconds to minutes.
To enforce structural changes on such timescales, biased MD simulation methods have been developed,
which aim at predicting free energies along a predefined reaction coordinate.
A problem with this type of simulation and analysis is that the resulting free energies only allow limited
inference of the unbiased systems's dynamics and nonequilibrium effects such as dissipation,
as fast degrees of freedom have been integrated out.
To allow such a coarse-graining of system dynamics, we have developed dissipation-corrected targeted molecular dynamics.
Combining the Jarzynski equality and the Markovian Langevin equation, we derive an expression for a dissipation correction
that can be calculated on-the-fly for nonequilibrium pulling simulations.
Our approach does not only result in the free energy profile, but in friction factors, as well,
the latter allowing insights into fast dynamics and fluctuations along the biasing coordinate as well as their molecular origin.
Furthermore, both quantities can serve as input for integration of a Langevin equation,
speeding up calculations of system dynamics by several orders of magnitude.
Combination with our approach of temperature boosted Langevin equation simulations finally allows to access timescales of up to minutes.
•
Targeted Molecular Dynamics Calculations of Free Energy Profiles Using a Nonequilibrium Friction Correction,
J. Chem. Theory Comput 12, 6175 (2018)
•
Multisecond ligand dissociation dynamics from atomistic simulations,
Nat. Commun. 11, 2918 (2020)
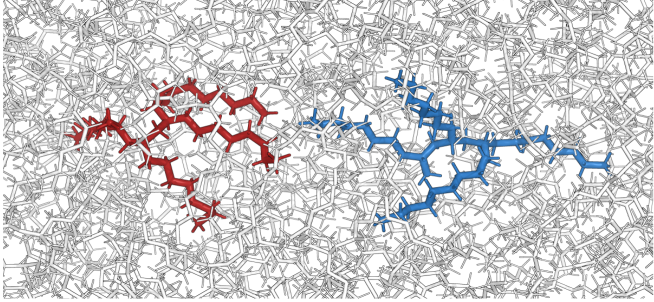
Molecular Origin of Driving-Dependent Friction in Fluids
The friction coefficient of fluids may become a function of the velocity at increased external driving. This non-Newtonian behavior is of general theoretical interest as well as of great practical importance, e.g., for the design of lubricants. Using dissipation-corrected targeted molecular dynamics, we pull apart two tagged liquid molecules in the presence of surrounding molecules to study this driving-dependence of the friction. Describing the process via a generalized Langevin equation, we show that velocity-dependent friction in fluids results from an intricate interplay of near-order structural effects and the non-Markovian behavior of the friction memory kernel. For complex fluids such as the model lubricant C40H82, the memory kernel exhibits a stretched-exponential long-time decay, which reflects the multitude of timescales of the system.
•
Molecular Origin of Driving-Dependent Friction in Fluids
JCTC 2022, 18, 5, 2816-2825
Computer simulations have become indispensable tools for exploring the dynamics of proteins.
However, as it is neither possible nor desirable to follow the motion of a complex molecule
along its 3N atomic coordinates, we need methods that can fully realize the potential of
the simulations.
One of the most important driving force for the modeling and analysis of MD simulation data
are Markov state models (MSM), which model the often bewildering dynamics of a protein by
memoryless, probabilistic jumps between its metastable configurations.
The key assumption behind Markov state models is that the dynamics of the system can be approximated
by a Markov chain meaning that the actual state of the system \(X\) does only depend on its predecessor
\( p( X_{T+1}| X_0, \dots, X_T) = p(X_{T+1}| X_{T}) \).
Therefore, a major challenge in Markov state modeling is to find such states \(X\) for which this
Markov property is fulfilled which requires different strategies to reduce the dimensionality of the
data while retaining the dynamical information.
• Perspective: Identification of collective variables and metastable states of protein dynamics,
J. Chem. Phys. 149, 150901 (2018)
• Dynamical coring of Markov state models,
J. Chem. Phys. 150, 094111 (2019)
A popular approach to analyze complex biomolecular systems is to define a low-dimensional
reaction coordinate which aims for an appropriate description of the essential collective
dynamics. Assuming a separation of time scales between those coordinates and the neglected
orthogonal degrees of freedom, the Markovian Langevin equation represents a powerful
framework to investigate and interpret the observed dynamics. We have developed a data-driven
Langevin equation (dLE) algorithm to estimate multidimensional
Langevin fields from MD data. Only based on local information, the dLE allows for the
interpretation of enhanced sampling data to predict long time dynamics like transition
times.
Based on dLE studies we proposed a practical method to construct an analytically defined
global model of structural dynamics. This approach employs an “empirical valence
bond”-type model which can cover multidimensional free energy landscapes as well as
an approximate description of the friction field estimated by the dLE. Adopting alanine
dipeptide and a three-dimensional model of heptaalanine as simple examples, the resulting
Langevin model reproduced the results of the underlying all-atom simulations.
For further applications, analytically defined models can be used to investigate the
dependence of the system on parameter changes and to predict the effect of site-selective
mutations on the dynamics.
• Global Langevin model of multidimensional biomolecular dynamics,
J. Chem. Phys. 145, 184114 (2016)
• Multidimensional Langevin Modeling of Nonoverdamped Dynamics,
Phys. Rev. Lett. 115,050602 (2015)
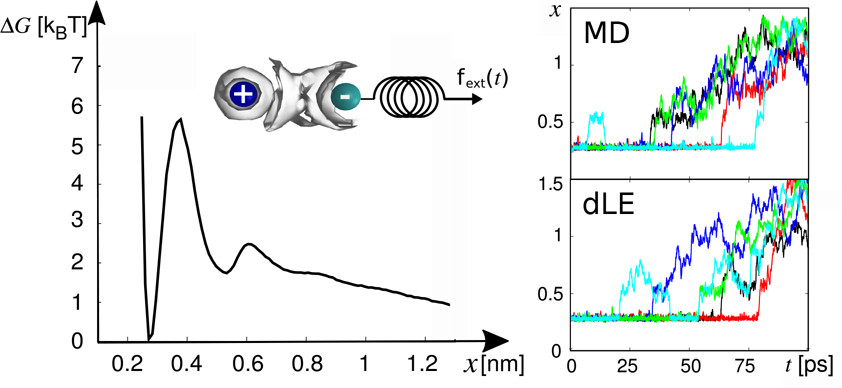
Data-driven Langevin modeling of nonequilibrium processes
Given nonstationary data from molecular dynamics simulations, a Markovian Langevin model is constructed that aims to reproduce the time evolution of the underlying process. While at equilibrium the free energy landscape is sampled, nonequilibrium processes can be associated with a biased energy landscape, which accounts for finite sampling effects and external driving. When the data-driven Langevin equation (dLE) approach [Phys. Rev. Lett. 2015, 115, 050602] is extended to the modeling of nonequilibrium processes, an efficient way to calculate multidimensional Langevin fields is outlined. The dLE is shown to correctly account for various nonequilibrium processes, including the enforced dissociation of sodium chloride in water, the pressure-jump induced nucleation of a liquid of hard spheres, and the conformational dynamics of a helical peptide sampled from nonstationary short trajectories.
•
Principal component analysis of nonequilibrium molecular dynamics simulations,
J. Phys. Chem. B 125, 8125 (2021)
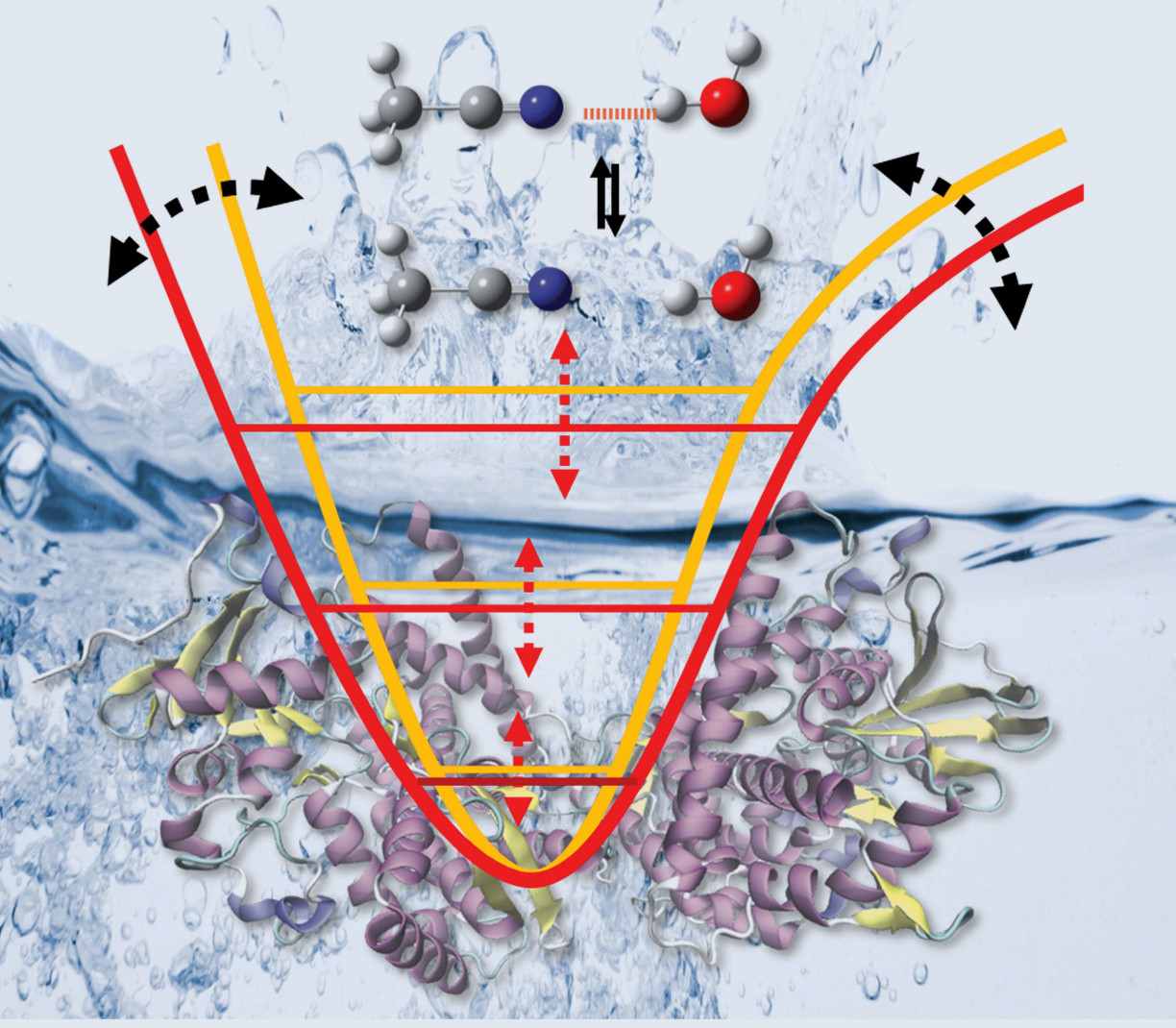
Modern techniques of infrared spectroscopy, in particular multidimensional and time-resolved version of it,
have opened the way to monitor secondary structure and dynamics of polypeptides and proteins.
In particular the amide I band, reflecting the backbone C=O stretching vibration,
has been found to yield structural information.
To obtain a first principles description of the amide I vibrational spectrum of peptides in aqueous solution,
we pursue a quantum-classical approach which consists of a classical molecular dynamics simulation of
the conformational distribution of the system, density functional theory calculations of the conformation-dependent
and solvent-induced frequency fluctuations, and a semiclassical description of the vibrational line shapes.
Vibrational Spectroscopic Map, Vibrational Spectroscopy, and Intermolecular Interaction
Real time observation of ultrafast peptide conformational dynamics: MD simulation vs. IR experiment
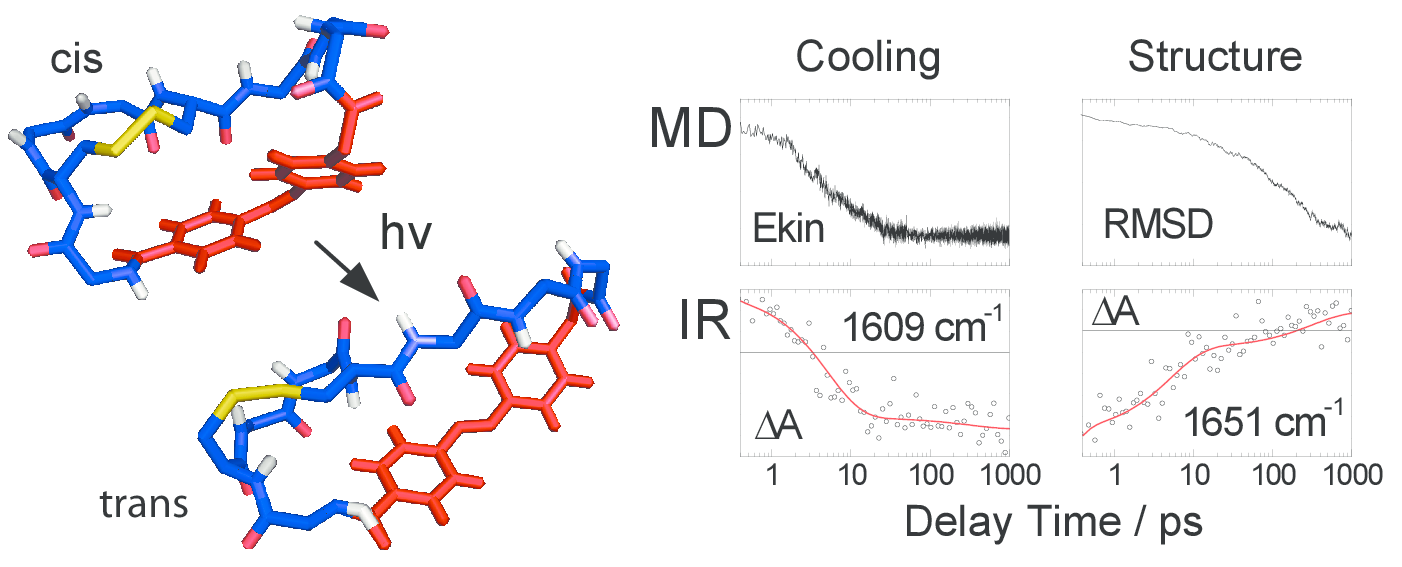
Employing nonequilibrium molecular dynamics (MD) simulations and transient infrared (IR) spectroscopy,
a joint theoretical/experimental study on a water-soluble photoswitchable octapeptide designed by
Renner et al. [Biopolymers 63, 382 (2002)] is presented.
The simulations predict the cooling of the hot photoproducts on a time scale of 7 ps and complex conformational
rearrangements ranging from a few picoseconds to several nanoseconds.
The experiments yield a dominant fast relaxation time of 5 ps, which is identified as the cooling time
of the peptide in water and also accounts for initial conformational changes of the system.
Moreover, a weaker component of 300 ps is found, which reflects the overall conformational relaxation of the system.
The virtues and the limitations of the joint MD/IR approach to describe biomolecular conformational rearrangements are discussed.
• Real Time Observation of Ultrafast Peptide Conformational Dynamics: Molecular Dynamics Simulation vs Infrared Experiment,
J. Phys. Chem. B 115, 13084 (2011)
Simulation of transient infrared spectra of a photoswitchable peptide
Ultrafast nonadiabatic photoreactions
Maximum Caliber
We show how to apply a general theoretical approach to nonequilibrium statistical mechanics, called Maximum Caliber,
originally suggested by E. T. Jaynes 30 years ago, to a problem of two-state dynamics. Maximum Caliber is a variational
principle for dynamics in the same spirit that Maximum Entropy is a variational principle for equilibrium statistical
mechanics. The central idea is to compute a dynamical partition function, a sum of weights over all microscopic paths,
rather than over microstates. We illustrate the method on the simple problem of two-state dynamics, A ↔ B, first
for a single particle, then for M articles. Maximum Caliber gives a unified framework for deriving all the relevant
dynamical properties, including the microtrajectories and all the moments of the time-dependent probability density.
While it can readily be used to derive the traditional master equation and the Langevin results, it goes beyond them in
also giving trajectory information. For example, we derive the Langevin noise distribution, rather than assuming it.
As a general approach to solving nonequilibrium statistical mechanics dynamical problems, Maximum Caliber has some advantages:
- It is partition-function-based, so we can draw insights from similarities to equilibrium statistical mechanics,
- it is trajectory-based, so it gives more dynamical information than population-based approaches like master equations; this is particularly important for few-particle and single-molecule systems,
- it gives an unambiguous way to relate flows to forces, which has traditionally posed challenges, and
- like Maximum Entropy, it may be useful for data analysis, specifically for time-dependent phenomena.
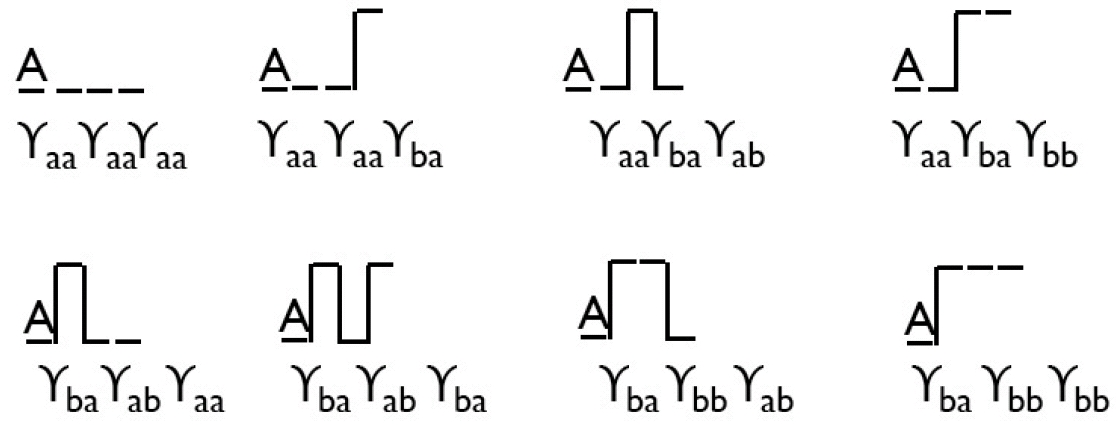
Graphical representation of the dynamical partition function of a two-state system, showing all possible trajectories for N=3 time steps for a system stating in A.
•
Maximum Caliber: A variational approach applied to two-state dynamics,
J. Chem. Phys. 128, 194102-194102-12 (2008)
•
Maximum caliber inference of nonequilibrium processes,
J. Chem. Phys. 133, 034119 (2010)
•
Inferring Transition Rates of Networks from Populations in Continuous-Time Markov Processes,
J. Chem. Theory Comput. 11, 5464 (2015)